Entamoeba histolytica research presented at the Molecular Parasitology Meeting
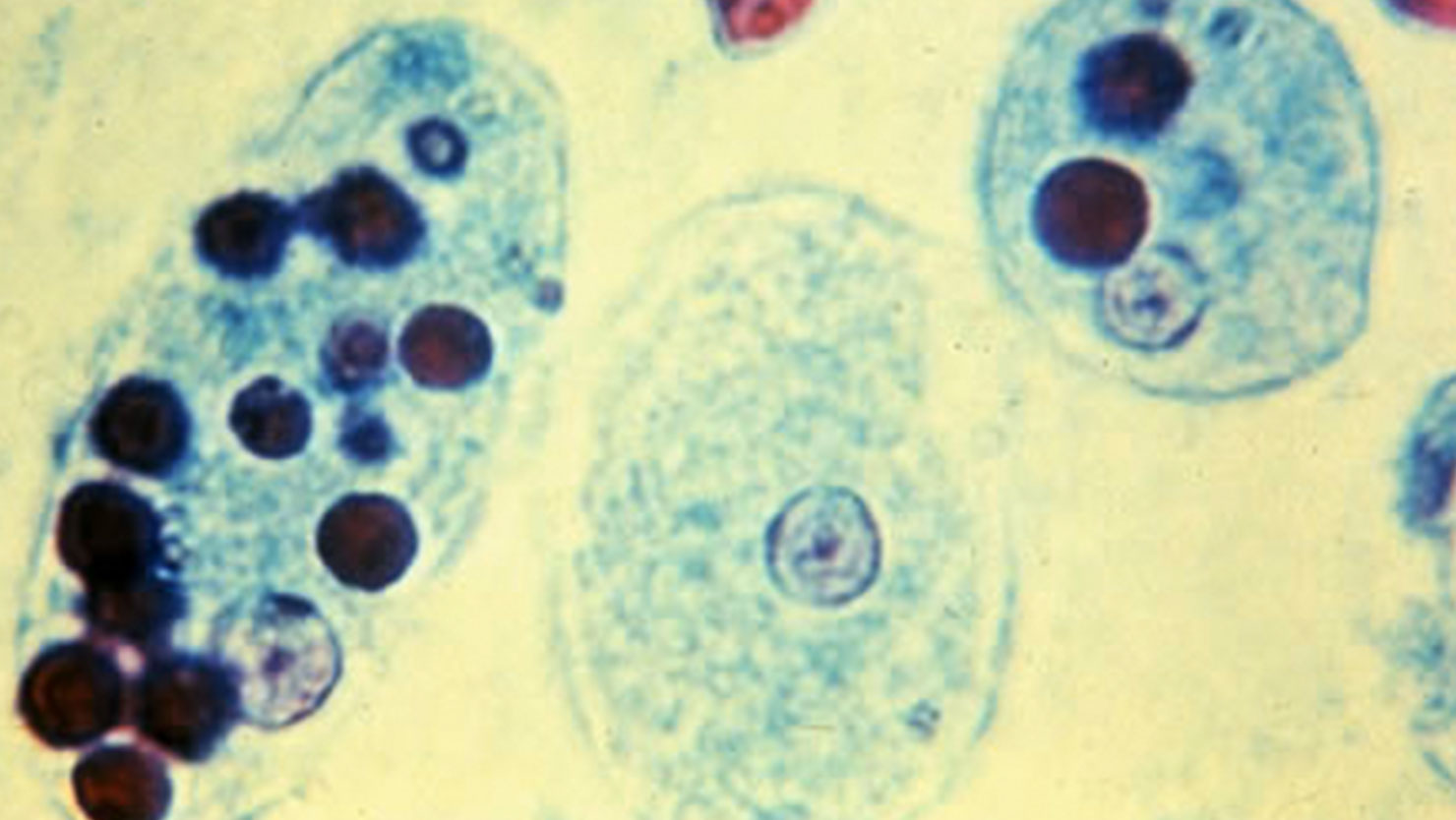
Entamoeba histolytica causes invasive intestinal and extraintestinal infections, known as amoebiasis, in about 50 million people and still remains a significant cause of human death in developing countries. However, for unknown reasons, fewer than 10% of E. histolytica infections are symptomatic (causing symptoms such as diarrhea, dysentery or liver abscess). The J. Craig Venter Institute is among the institutions awarded the NIAID Genome Sequencing Centers for Infectious Diseases (GSCID) contracts to provide high-quality genome sequencing and high-throughput genotyping of NIAID Category A-C priority pathogens.
A GSCID project led at JCVI by Dr. Elisabet Caler includes performing whole-genome sequencing of Entamoeba phenotypic variants from symptomatic, asymptomatic and liver abscess-causing strains chosen to include a range of clinical manifestations and taken from human cases, as well as strains grown under different conditions. Our objective is to develop a genome-wide landscape of Entamoeba diversity to understand how sequence variations in the parasite relate to pathogenicity (ability to cause disease) and clinical outcome.
The Molecular Parasitology Meeting held at the Woods Hole Oceanographic Institution, Woods Hole, MA last week provided a window into the exciting science of Parasitology. The keynote speaker, Fotis Kafatos, spoke on "Major Challenges to Global Health in the Tropics and Beyond--Insect Vectors of Malaria and Other Parasitic or Viral Diseases." Dr. Kafatos stressed that a multi-pronged approach to the control of malaria is necessary to prevent the devastating loss of life that malaria causes.

The many excellent papers and posters provided an overview of the field, including Plasmodium falciparum, Toxoplasma gondii, the trypanosomes, Giardia lamblia, Trichomonas vaginalis, Entamoeba histolytica, Schistosoma species, Babesia bovis, and associated vectors. Topics spanned basic biology, drug design, sequencing and host-pathogen interactions.
I presented an overview of the Entamoeba sequencing project at the meeting. Discussions as a result of the presentation included questions about the details of sequencing and handling the next-generation sequencing data. We had animated discussions about methods for assembly of the DNA sequences, including reference-guided vs de novo assembly. Many attendees were impressed with JCVI’s open-source METAREP metagenomic tool (J. Goll, et al., Bioinformatics 2010). Determination of the best methods for the analysis of differences in the clinical isolates generated much discussion. Entamoeba researchers see the sequences as a great resource and are looking forward to being able to mine the data. One, from India, was very excited that he was going to have about 15 times the resources he has had in the past, since he has had only had one genome to mine up until now.
The Molecular Parasitology Meeting was an excellent venue for scientific exchange. The Entamoeba histolytica GSCID project will help us understand the pathogenicity of Entamoeba histolytica, and has the potential to save lives in developing countries.