SARS-CoV-2 Mutation Tracking
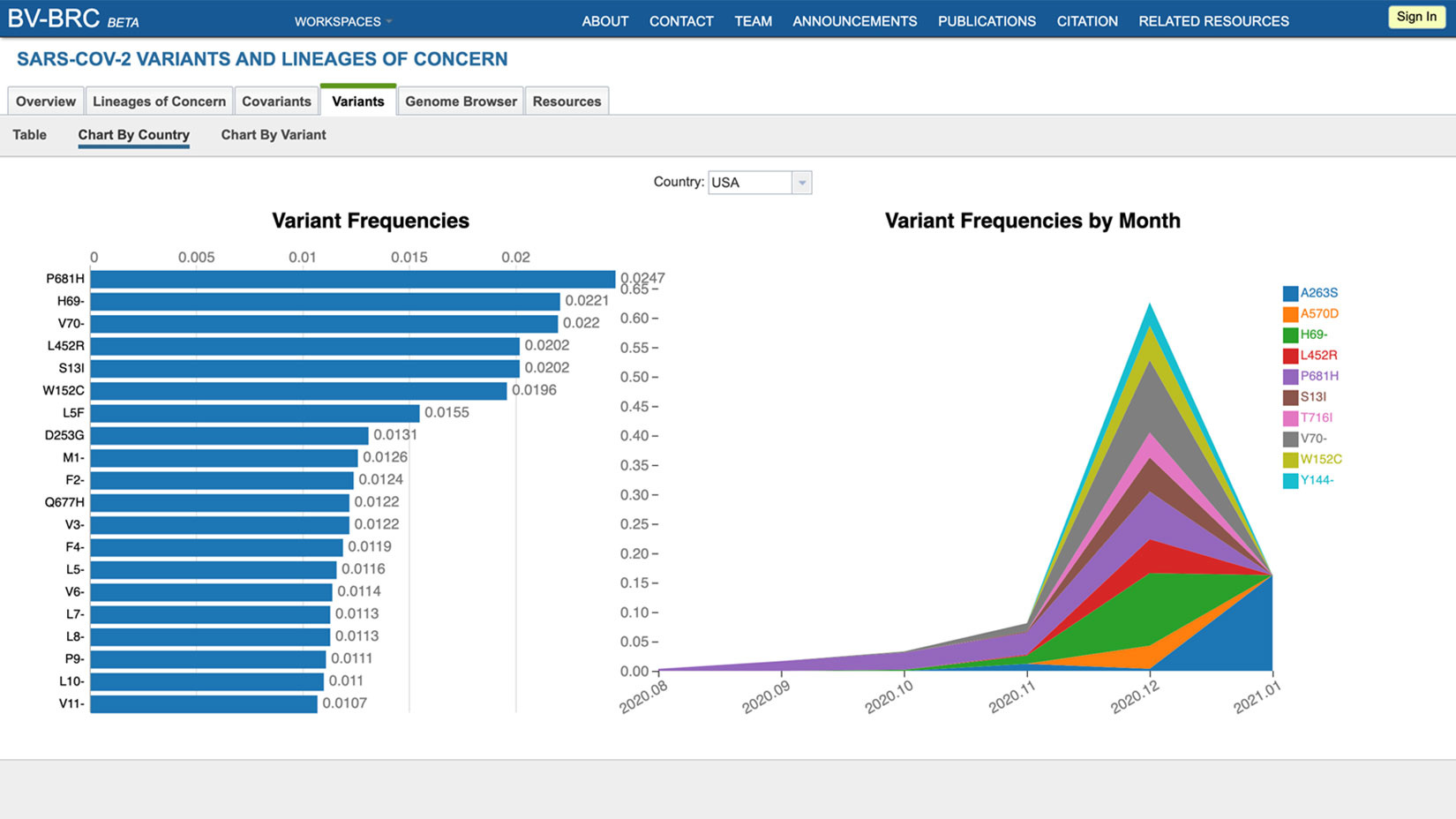
The Bacterial Viral Bioinformatic Resource Center (BV-BRC) is proud to introduce a new resource with the goal of providing live tracking of SARS-CoV-2 mutations. This real-time resource will provide regular reports focused on “Variants and Lineages of Concern” (VoCs/LoCs), and will serve as an early warning system for variants that are increasing in frequency in specific geographical locations.
In addition to performing risk assessments on variants to identify candidate VoCs/LoCs, the system also leverages a growing knowledgebase of sequence features, including protein domains, functional regions, and immune epitopes to relate genome changes to functional consequences.
Several dashboards are available to view sequence prevalence by countries and regions over time. The BV-BRC also provides a genome browser, protein structure viewer, and phylogenetic trees in order to facilitate a better understanding of how these changes in the viral genome affect viral phenotypes and disease dynamics.
Users can access the new BV-BRC SARS-CoV-2 Variants and Lineages of Concern resource at https://bv-brc.org/view/VariantLineage/#view_tab=overview. This resourse is being developed by scientists at J. Craig Venter Institute (JCVI), University of Chicago, University of Virginia, and Argonne National Lab.