Virtual Comparative Metagenomics
We have created an open virtualization format (OVF) package of JCVI's Metagenomics Reports (METAREP)- a high performance comparative metagenomics analysis tool. The software runs on a web server, retrieves data from two different database systems and uses R for statistical analysis. The new OVF package bundles all these 3rd party tools and is configured to run out of the box in a virtual machine.
To run a virtual version of METAREP on your machine, follow these steps
- download the METAREP OVF package from our ftp site [download] .
- unzipp the OVF package
- download and install Oracle's Virtual Box, a OVF compatible virtualization software [download]
- Start Virtual Box
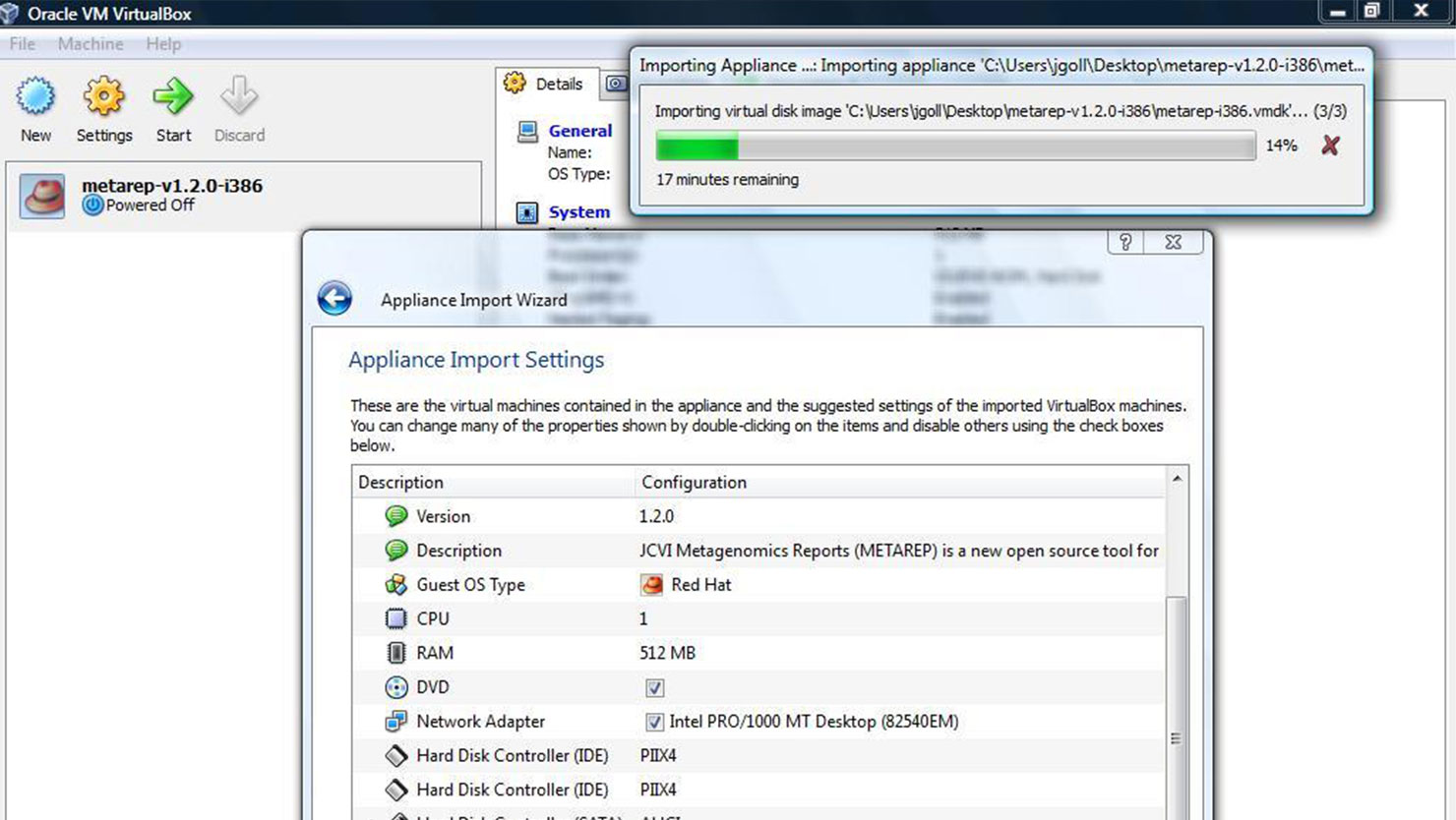
- Click File/Import Appliance and select the OVF file.
- Adjust RAM/CPU usage using the Appliance Import Wizard (see image)
- Start VM
- Double-Click on the METAREP firefox link on the VM desktop
- Log into METAREP with username=admin and password=admin
This virtual machine appliance is the first step in developing a fully cloud-enabled analysis platform where users can easily launch the application wherever is most convenient: on their personal desktop or in the cloud where they can scale-out the appliance to suite their needs.
Future virtual machine images will be certified to run on other virtualization software platforms. Stay tuned.
If you like to learn more about METAREP and talk to the developers, join us at Lucene Revolution Conference in Boston (October 7-8, 2010). We will present a lightning talk about METAREP the first day of the conference 5pm (see agenda).
Links: