First Minimal Synthetic Bacterial Cell
Researchers from the J. Craig Venter Institute (JCVI) and Synthetic Genomics, Inc. (SGI) have accomplished the next feat in synthetic biology research—the design and construction of the first minimal synthetic bacterial cell, JCVI-syn3.0.
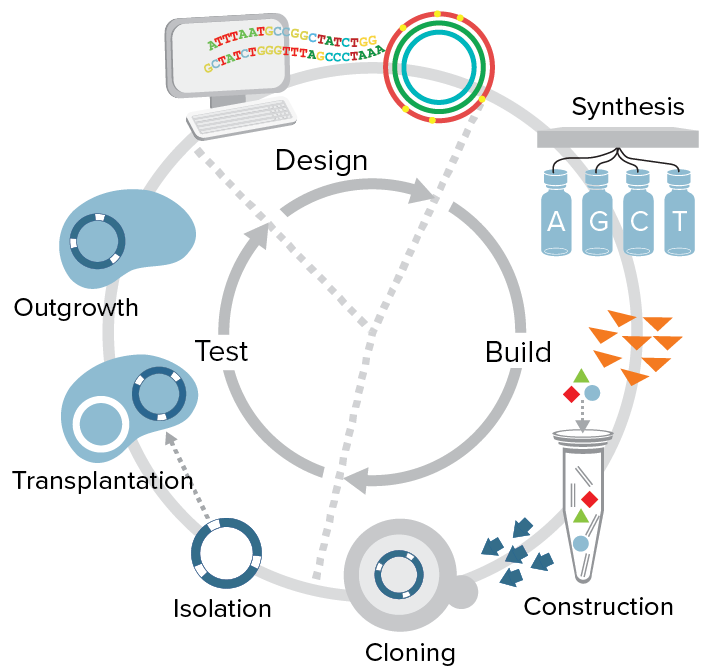
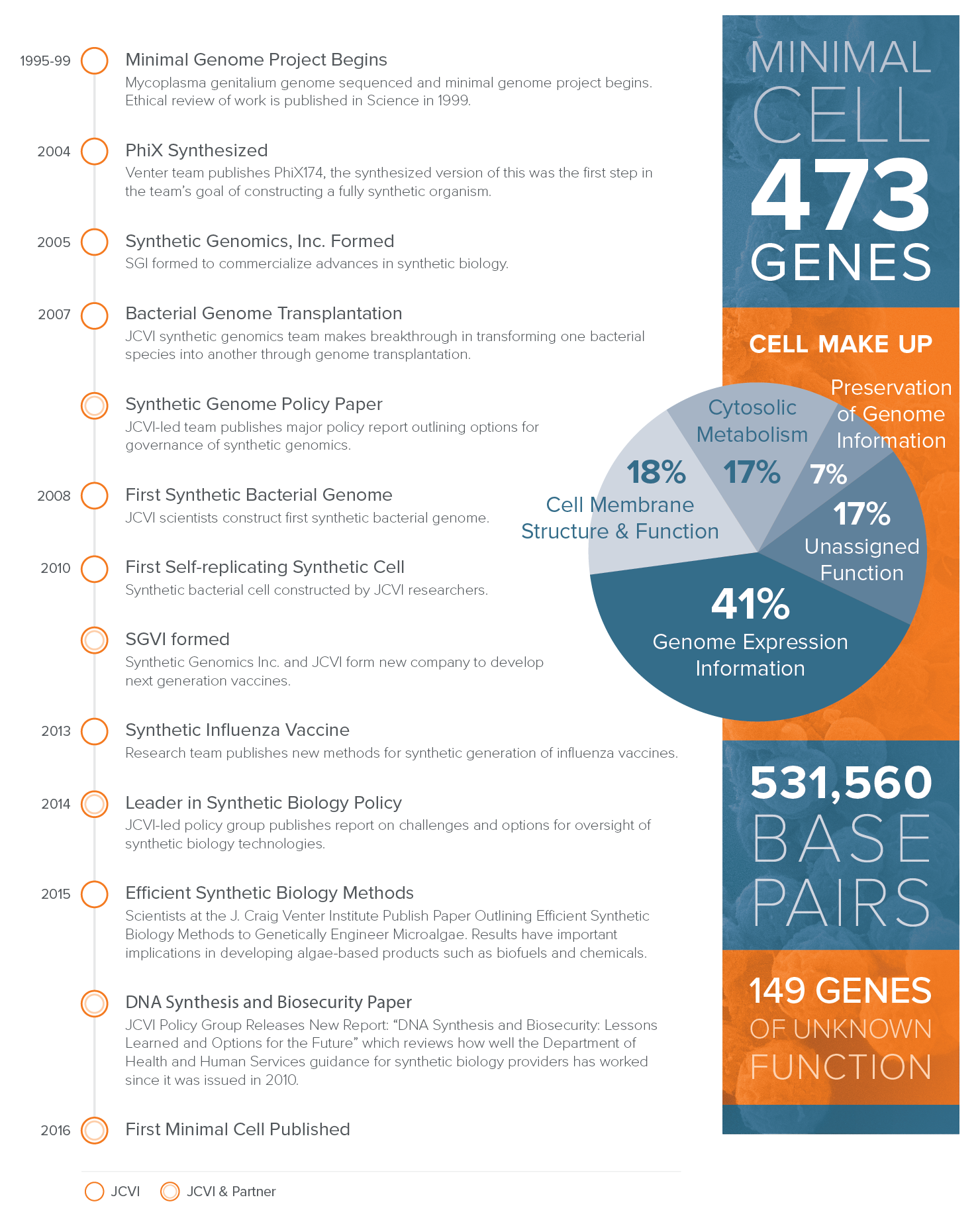
Using the first synthetic cell, Mycoplasma mycoides JCVI-syn1.0 (built by this same team in 2010), JCVI-syn3.0 was developed through a design, build, and test (DBT) process using genes from JCVI-syn1.0. The new minimal synthetic cell contains only 531,000 base pairs and just 473 genes making it the smallest genome of any self-replicating organism.
 A paper describing this research is published in the March 25 print version of the journal, Science by lead author Clyde A. Hutchison, III, PhD, senior author J. Craig Venter, PhD, and senior team of Hamilton O. Smith, MD, Daniel G. Gibson, PhD, and John I. Glass, PhD.
A paper describing this research is published in the March 25 print version of the journal, Science by lead author Clyde A. Hutchison, III, PhD, senior author J. Craig Venter, PhD, and senior team of Hamilton O. Smith, MD, Daniel G. Gibson, PhD, and John I. Glass, PhD.
Writing Biological Code
A biological cell is very much like a computer—the genome is the software that encodes the instructions of the cell and the cellular machinery is the hardware that interprets and runs the genome software. Major advances in DNA technologies have made it possible for biologists to now behave as software engineers and rewrite entire genomes to program new biological operating systems.
Project Goals
A major goal in synthetic biology is to have the capacity to predictably design and build DNA that produces a cell with new and improved biological functions that do not already exist in nature. Significant advances have been made in DNA design at the gene and pathway level and in engineering bacteriophage genomes. But, even with all the advances that have been made in genomics and synthetic biology, there is still not a single self-replicating cell in which we understand the function of every one of its genes. Toward this goal, the JCVI/SGI team has been working to understand the gene content of a minimal cell—a cell that has only the machinery necessary for independent life.
Since the generation of the first synthetic cell in 2010, the team found ways to drastically speed up the process of building cells from the bottom up. They developed new tools and semi-automated processes for genome synthesis, including more rapid, more accurate, and more robust methods for going from oligonucleotides (small pieces of DNA) to whole chromosomes. Over the past 10 years whole bacterial chromosome assembly has gone from impossible, to possible in years, to months and now to just weeks with these new methods, which are made available to scientists in this manuscript.
A major outcome of this minimal cell program has been new tools and semi-automated processes for whole genome synthesis. Many of these synthetic biology tools and services are commercially available through SGI-DNA.
Milestones in JCVI Synthetic Biology Research and Policy
Funding
This work was funded by SGI, the JCVI endowment and the Defense Advanced Research Projects Agency’s Living Foundries program, HR0011-12-C-0063.
Principal Investigator
Note on Data
The JCVI-syn3.0 sequence has been deposited in Genbank under the accession number CP014940. You can access the supplementary material for the paper by clicking on the link at the end of the text. Note that in addition to Materials and Methods, Supplementary Figures, Supplementary Tables, and References, there is also a link to an Excel Spreadsheet named Database S1. This spreadsheet gives detailed information about the genes of JCVI-syn3.0. The coordinates of each gene on the M. mycoides JCVI-syn1.0 sequence are listed (Genbank accession number CP002027). There is also information about gene annotation, and a column listing the complete amino acid sequence of each protein gene product.