Discriminative Machine Learning for Blood Cancer Precision Diagnostics
Diagnosis of blood cancer usually requires accurate identification of cancer cell populations from blood and bone marrow samples. Flow cytometry (FCM) is a primary diagnostic assay routinely used in clinical practice for leukemia diagnosis. The assay workflows consist of multiple manual analysis steps performed by technicians, followed by interpretation by hematopathologists. Challenges to this process include technical variability in the manual analysis, difficulty in identification of the atypical leukemic cells, and the growing number of antigens used for diagnosis.
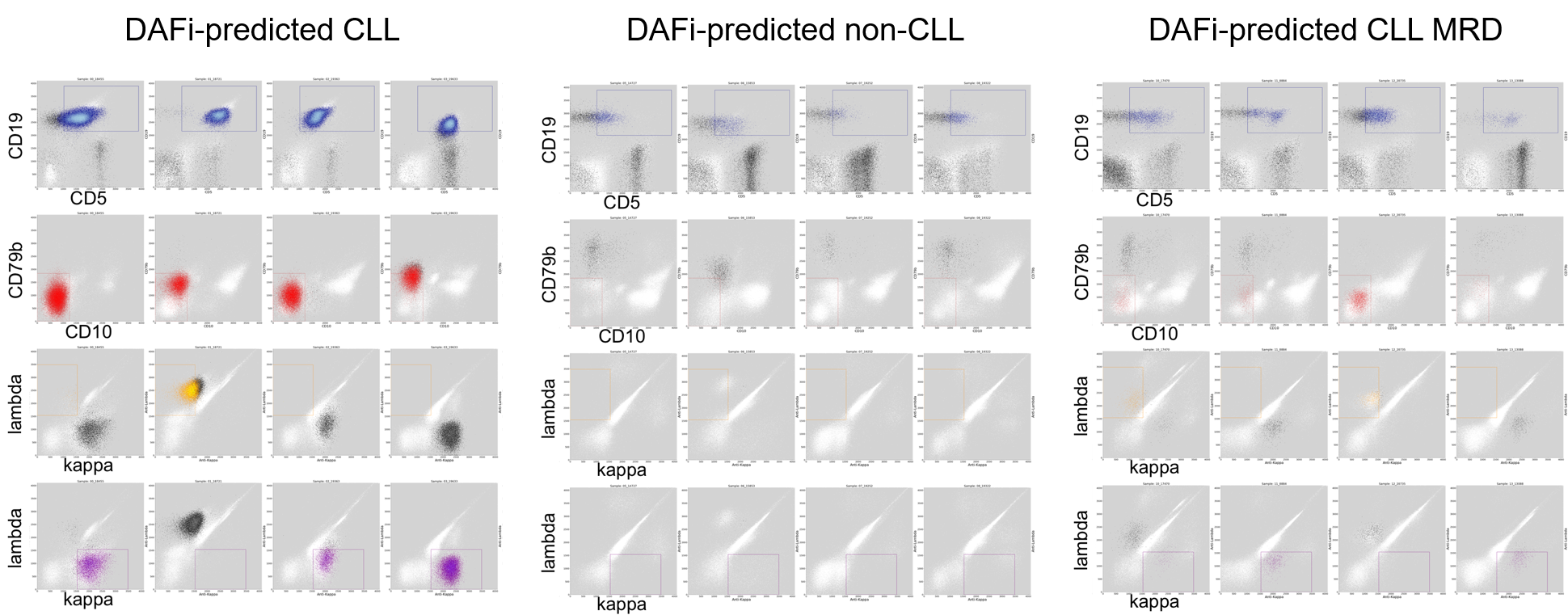
Instead of conducting ad hoc analysis of individual patient samples, we developed a suite of machine learning methods to leverage preexisting clinical FCM samples for improving the precision identification of leukemic cells. Our discriminative learning method optimizes both cell population identification and sample classification simultaneously, making the “black box” machine learning classification interpretable with results recognizable to hematopathologists.
Collaborating with researchers at Stanford, UC Irvine, and UC San Diego, we lead a 5-year project for developing a web-based computational infrastructure – FlowGate – to improve the accessibility and usability of the cutting-edge cytometry data analytical approaches for both translational research and clinical diagnosis. The back-end infrastructure is built upon a large cluster computer at San Diego Supercomputer Center with 1,944 compute nodes to support web-based interactive analytics and visualization across samples.
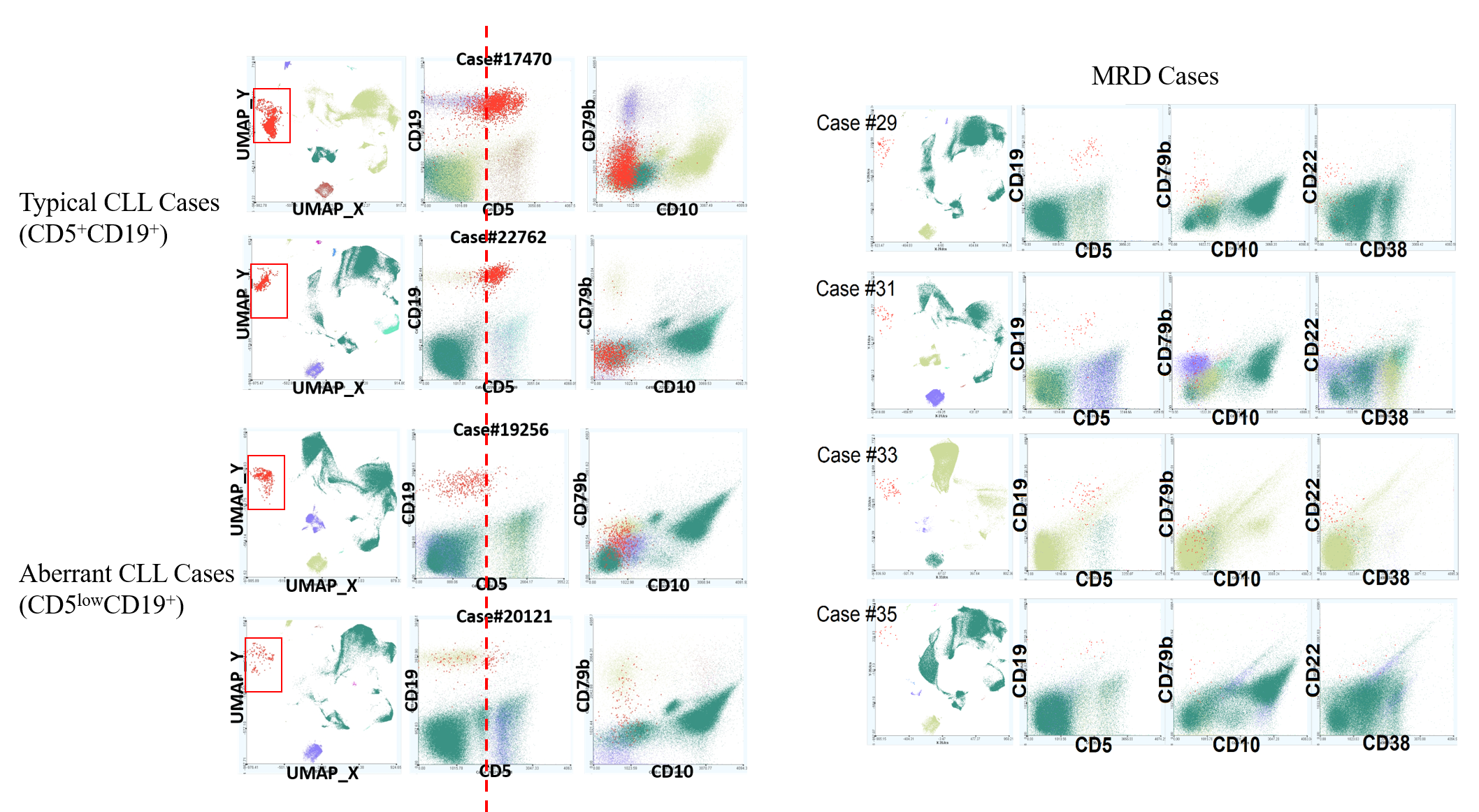
Our experiments showed that both typical and atypical types of chronic lymphocytic leukemia (CLL) cells can be clearly captured using our computational approach. The approach is general and potentially applicable to other types of blood cancers. We are working with collaborators from diagnostics labs at Stanford, CHLA of USC, University of Washington, and UC San Diego to extend and apply the machine learning approach to diagnosis of more types of blood cancers including acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), and multiple myeloma (MM). Our goal is to improve and utilize machine intelligence for elucidating cancer heterogeneity and disease endotypes for supporting cancer precision medicine.
Key Findings
- Automated gating analysis reduces human bias in current manual identification of leukemic cells
- We developed a novel discriminative learning approach that can optimize gating locations and sample classification simultaneously
- Non-linear embedding dimensionality reduction can be used with automated gating analysis to improve identification of atypical CLL cells that otherwise could not be identified