Candida Profiling and Gene Expression During Host Infection
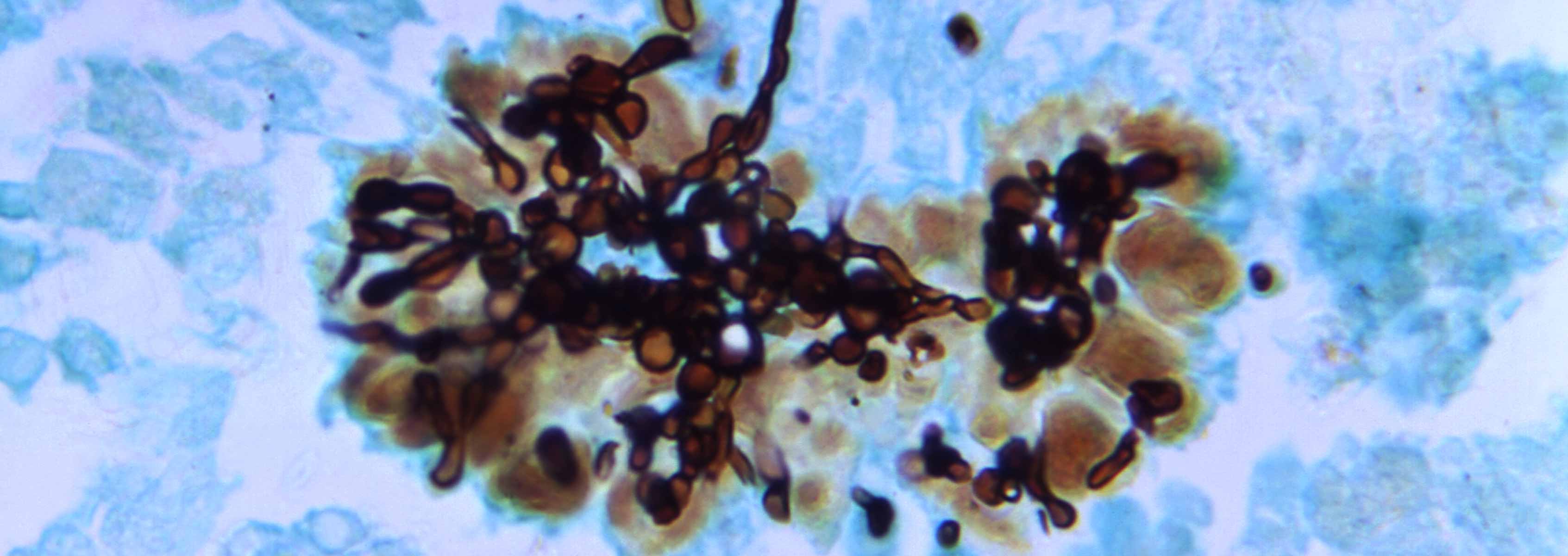
One of the most common cause of human fungal infections is Candida species. We have several studies on going to determine the molecular mechanisms of interactions between Candida species and host cells. Bill Nierman at JCVI with Hong Nguyen, Neil Clancy and Paschalis Vergidis at The University of Pittsburgh, Department of Medicine and Scott Filler at Harbor-UCLA Medical Center have collaborated to define the genomic characteristics and pathogenesis of Candida glabrata and Candida albicans during mouse and human Intra-Abdominal infections.
To better characterize C. glabrata, JCVI performed RNA sequencing (RNAseq) analysis from mouse peritoneal fluid and abscesses from a time course experiment to assess gene expression changes over time. In a similar project, we performed mouse infection studies and determined the effect of the fungal pathogen C. albicans on host transcriptome, especially when it is introduced along with sterilized stool. At JCVI, we processed peritoneal fluid and abscess samples for RNAseq, and determined the transcript abundance for each C. albicans and mouse transcripts. Results of these analyses will reveal molecular details about the host response to C. albicans. The Nierman team was also involved in RNAseq analysis with C. albicans mutants in the absence and presence of Caspofungin to determine time course expression in response to Caspofungin with the aim of producing a drug exposure profile.
In addition, tThe Nierman team along with Drs. Nguyen and Clancy used RNAseq to profile C. albicans transcription within biliary fluid from a patient with cholangitis; samples were collected before and after treatment with fluconazole and biliary tract drainage. C. albicans transcriptomes at the infection site distinguished treated from untreated cholangitis. After treatment, 1131 C. albicans genes were differentially expressed in biliary fluid. Up-regulated genes were enriched in hyphal growth, cell wall organization, adhesion, oxidation reduction, biofilm, and fatty acid and ergosterol biosynthesis. This is the first study to define Candida global gene expression during deep-seated human infection. Successful treatment of cholangitis induced C. albicans genes involved in fluconazole responses and pathogenesis.
Our collaborations with and Scott Filler at Harbor-UCLA Medical Center focused on microbiome signatures that are associated with C. albicans colonization during antibiotic exposure. Specifically, we used a mouse model to investigate the risk factors during antibiotic therapy for opportunistic, gastrointestinal (GI) colonization with C. albicans, a commensal fungus in the human GI tract. C. albicans does not normally colonize mice unless the resident GI flora is perturbed by antibiotics. Therefore, we exposed mice to a single challenge with C. albicans 7 days after initiating antibiotic treatments that spanned a total of 21 days. Our initial observations indicated that some antibiotics induced a persistently higher level of C. albicans colonization compared to others. We sought further insights into these differential patterns of colonization by identifying the underlying antibiotic-induced perturbations in the bacterial and fungal GI microbiome, and the host immune factors. Hence, we employed a regression framework to explain the level of C. albicans colonization using microbiome variables (bacterial 16S and fungal ITS), immune factors and experimental conditions as covariables.
Funding
National Institutes of Health (NIH) funding provided through a subcontract from the University of Pittsburgh.